5. FAQ
5.FAQ.RmdFAQ
Below is an FAQ
Hardware recommendations
Below are the specs of the computers we use for processing our datasets.
- RAM: 64GB
- CPU: Intel(R) Core(TM) i9-10900
We recommend a minimum of 32 GB of RAM because the image processing scripts can be intensive.
How long should segmentation take?
It depends on the specs of your computer and the size and specs of your images. Since our images are somewhat high resolution, and our image segmentation macro segments in 3D not 2D, it can be relatively computationally intensive. However, it is easy to process an equivalent dataset of ~150 images over a weekend using a computer with the recommended hardware specs.
What are some common errors I might encounter either using SMARTR or in wholebrain?
R out of bounds error during registration or mapping
The most common error encountered during registration or mapping is an “out of bounds error” where the transformation index is out of bound.
This is largely due when the detected boundary (blue) around the border of the tissue is poor a poor fit. Wholebrain fits 32 correspondance points around the detected border to generate a first pass registration, and if the boundary touches the borders of the images, or goes outside the borders of the image, it throws this error.
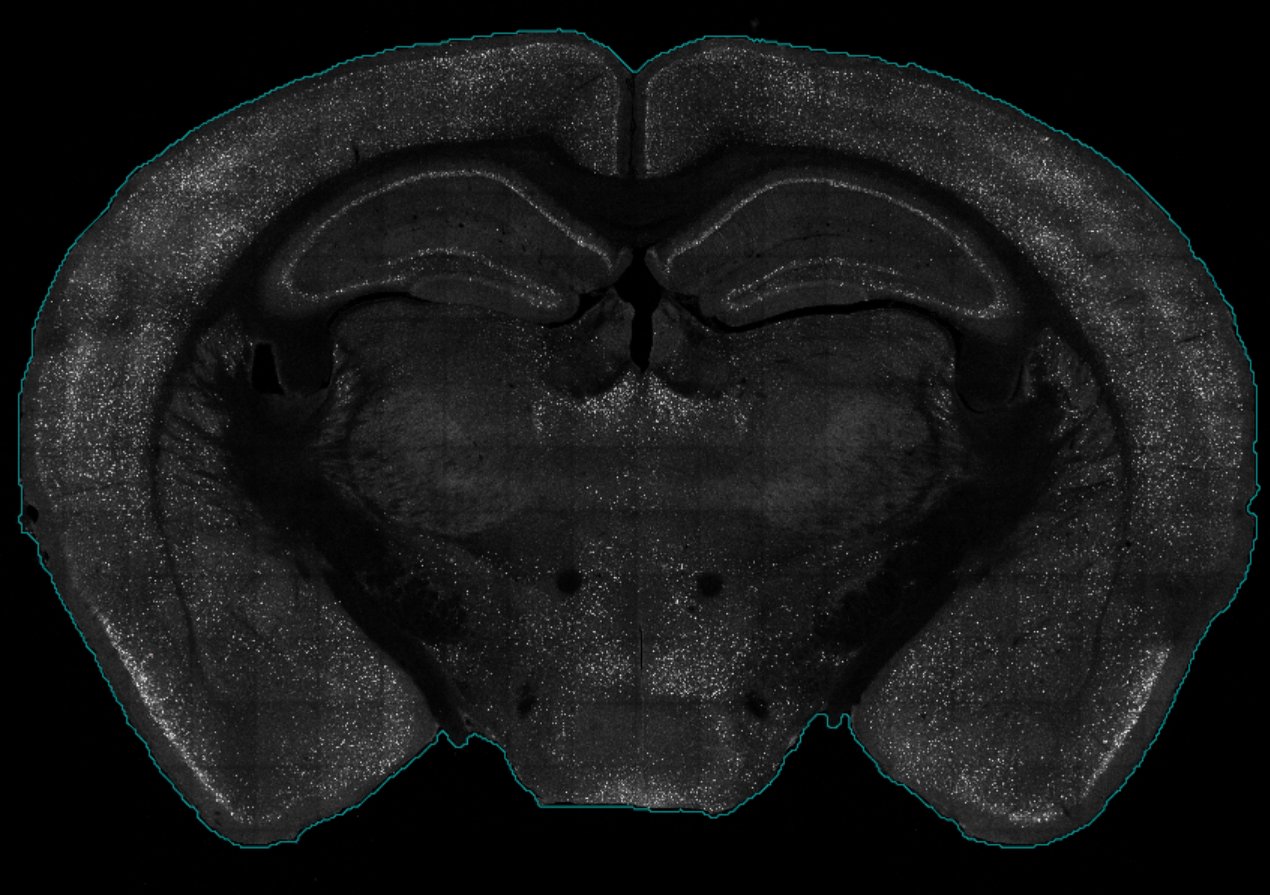
Not choosing the optimal brain.threshold
Since the detection of the tissue border depends on various factors, such as image brightness, it is possible that the poor border detection is due to an poor brain.threshold value. Please check out section 3.1 Detecting brain contours of the Tutorial page to see how to modify the brain threshold.
I can’t get a good first contour around my brains
Unfortunately, the contour detection can be buggy and inconsistent. Please check out section 3.1 Detecting brain contours of the tutorial page to see how to quickly and interactively adjust the brain.threshold to try out a wide range of values. If no optimal threshold is found, we recommend increasing the brain.threshold value until the contours detected are smaller than the actual tissue boundaries. Then proceed with registration. The first pass registration will be inaccurate. You can quickly manually correct the correspondance points using the interactive console interface.
The contours of my actual tissue are not contained in the borders of the image
If the brain contours in the image used for registration, i.e. the
MAX_name.tif in the 1. Imaging, segmentation, and
preprocessing section, are touching the 4 borders of the image, this may
throw an out of bounds error. These images should either be manually
edited in ImageJ to omit brain contours intersecting the image border,
or the original .tif image should be padded with a small
border of black pixels.
Not choosing the right resize parameter
The other scenario that may result in this error is when the size of the atlas overlay during registration is larger than the tissue boundary. I’m not sure why this error occurs, but this is corrected by changing the size of the atlas overlay.

Following boundary detection, change the size of the atlas overlay by
modifying the resize parameter of the filter. The optimal
range will depend on your imaging resolution
# Optimal value range for our imaging resolution is 0.8-0.12
# Larger resize values result in smaller overlays
filter$resize <- 0.1 None of the above fixed my issue.
As SMARTR is a wrapper around registration and mapping functions in wholebrain, we also recommend checking out the wholebrain github for more detailed questions.
I have damage to my brain and I want to exclude it from my analysis
The ability to process only subsets of regions are built-in to the SMARTR package. The sections below are organized in order of increasing damage severity.
I only want to exclude a subset of regions
This capability is part of the exclude_anatomy()
function and is accounted for using the
exclude_right_regions and exclude_left_regions
parameters.
You can enter an character vector of region abbreviations from the Allen
Brain Ontology to either of these parameters to add additional
regions to exclude. Note that any regions acronyms maintain the
heirarchical nature of the ontology, so if you wanted to exclude the
entire thalamus, instead of listing out every subregion, you can just
enter TH.
Additionally, by default layer 1 of cortical layer, ventricles, and
fiber tracts are already excluded. This is because these are the default
arguments for the left_regions_excluded and
right_regions_excluded parameters when creating a slice
object. You can set your own default regions to exclude, in addition to
those further excluded using exclude_anatomy.
I have regions torn and I only want to process one hemisphere?
If one hemisphere is totally ripped or beyond salvage, you can simply
set the hemisphere parameter when you create a slice object
to either left or right to mark this slice for
analysis of only the left or right hemisphere. The default value of
NULL means both hemispheres will be processed. Later when
exclude_anatomy() is run, simply omit the unwanted
contralateral hemisphere by setting exclude_hemisphere to
TRUE.
You can take advantage of this feature if as well if there is laterality in the sectioning, and the left hemisphere needs to be treated as a separate hemisphere and slice object from the right. Thus, separate atlas coordinates can be assigned to either hemisphere.
I really only want to process one or two regions per slice
This is an unusual case, but typically happens when a section is
almost completely ripped but there are one or two regions that remain
intact and can still be processed. To solely keep analyzing these
regions per hemisphere (and omit all other regions), enter the regions
to keep as the include_left_regions and
include_right_regions during the
exclude_anatomy() step. We do, however, recommend against
processing these sections if they can be avoided to ensure only high
quality sections in the dataset.
Alternatively, this can be used if you are intentionally analyzing
one or two regions using the pipeline, but still taking advantage of the
atlas overlay capabilities. You can optimize registration for these few
regions without regard for registering the whole plate, and use
include_right_regions and include_left_regions
to only process the regions being analyzed. To set these regions
beforehand, you can also enter these as parameters when making a
slice object.
I get a graphics issue
Could not find function “quartz”
If you get the following error:
Error in quartz(width = plate.width * 11.3, height = 7.9) :
could not find function "quartz"The function quartz is calling a graphics device driver for the macOS
system, so this error occurs when running the pipeline in Windows or
Linux. Calling quartz is built into some internals of
wholebrain functions. One work around is by running the lines below
every time you load the SMARTR library if you are running Windows or
Linux (This can only be done on machines/accounts that have access to an
X server.).
quartz <- x11