Welcome to the SMARTR package!
SMARTR is a package designed for the high-throughput mapping and analysis of dual-labelled ensemble datasets. Genetic tagging strategies such as the ArcCreERT2 mouse line allow for investigation of neural ensembles underlying behavior during multiple time points.
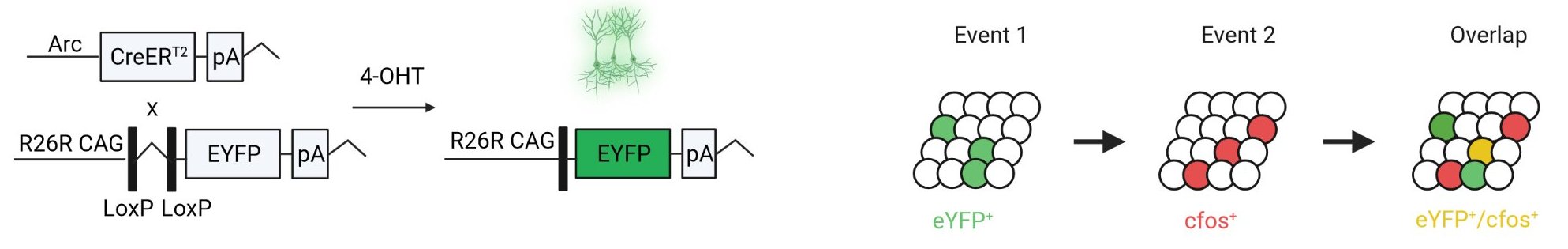
However, few tools exist to map dual-labelled ensembles, as well as their overlap across the brain in traditional coronal sections to a standardized atlas space. The SMARTR package facilitates this by bridging an optimized approach for cell-segmentation and colocalization that was developed in FIJI/ImageJ with registration and mapping capabilities from the wholebrain and SMART packages.
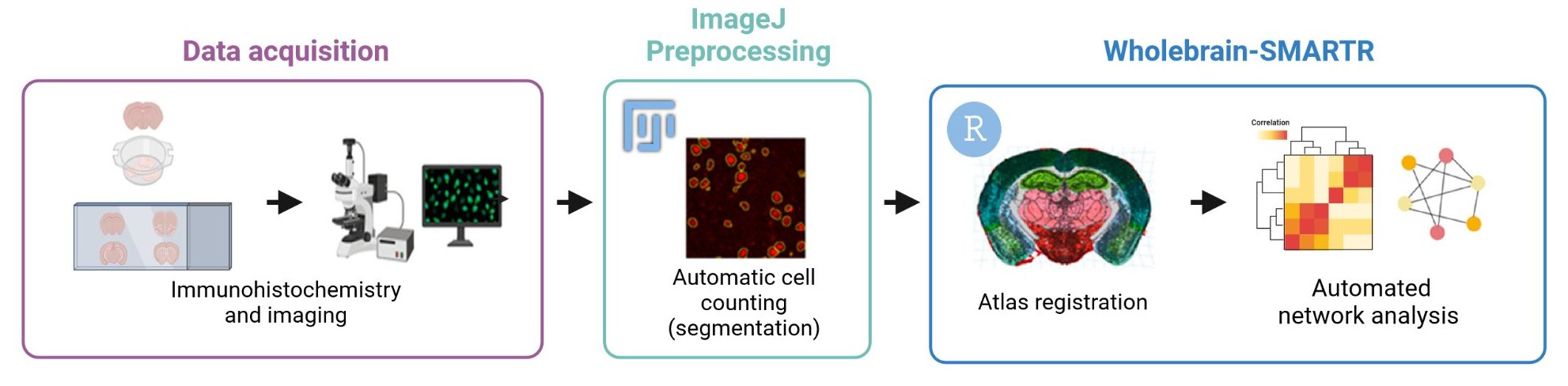
Moreover, SMARTR provides a streamlined API for storing metadata related to imaging, animal subject, and experimental parameters and groupings. Data management is intrinsically built into the package, which helps automate the process of combining ensemble datasets across multiple images, animals, and experimental groupings.
Finally, SMARTR provides a set of built-in analysis and visualization functions to conduct network analysis for each ensemble dataset for immediate downstream analysis. SMARTR is designed to facilitate dual-ensemble brain mapping projects by lowering the technical barrier for registration, segmentation, and statistical analysis.
Check out the Get Started page for installation instructions!
Publication citation
Use of the SMARTR package has been published on eLife as a reviewed preprint. If you are using SMARTR for academic purposes, please use the following citation:
Contact
Answers to common questions may be found in the FAQ page. Please look there first to troubleshoot any issues. If questions are not otherwise answered through the FAQ or through the tutorial, feel free to contact me at mj2947@cumc.columbia.edu for further clarification.